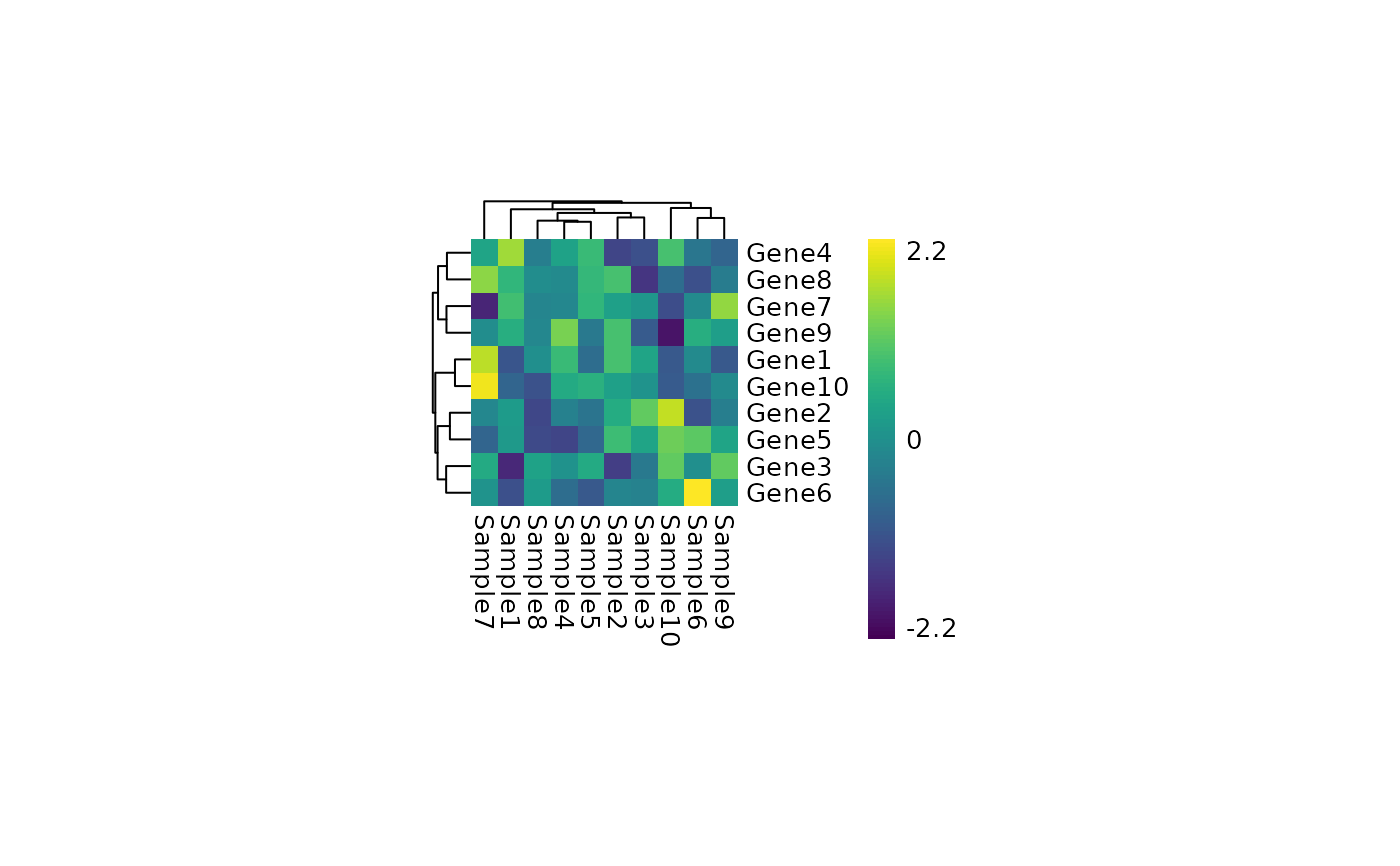create and save a nice pheatmap in the folder heatmap using color breaks
Arguments
- matrix
numeric matrix of the values to be plotted
- scale
should the values be centered and scaled in row, column direction? Allowed:
row,column,none(default:none)- height
height of output plot (default: number of rows divided by three)
- width
width of output plot (default: number of columns divided by two)
- cellwidth
individual cell width (default: 10)
- cellheight
individual cell height (default: 10)
- treeheight_row
height of a tree for rows (default: 10)
- treeheight_col
height of a tree for columns (default: 10)
- fontsize
fontsize (default: 10)
- cluster_rows
cluster rows? (default: true)
- cluster_cols
cluster columns? (default: true)
- annotation_row
data frame that contains the annotations. Rows in the data and in the annotation are matched using row names. (default: NA)
- dir_output
directory to save the output plot (default: ".")